
Model Deployment
John Harrold & Anson K Abraham
2026-01-07
Source:vignettes/Deployment.Rmd
Deployment.RmdIntroduction
Once a model has been developed it may be useful to provide an easy way to run simulations or automate specific analyses. Do to that, a ubiquity provides a ShinyApp that allows the model to be run from an easy to use interface that is highly customizable. This App can be run locally (useful during meetings to answer quick questions) or deployed on a Shiny server to allow others access to the model.
To demonstrate this interface we can begin with the PK model of mAbs in humans (Davda etal. mAbs, 6(4), 1094-1102) :
library(ubiquity)
system_new(file_name = "system.txt", system_file = "mab_pk", overwrite = TRUE)
cfg = build_system(system_file = "system.txt")Use ?system_new to see a list of the available system
file examples. After building any system you can then create the
ShinyApp template:
system_fetch_template(cfg, template = "ShinyApp", overwrite = TRUE)This should create the following three files:
-
ubiquity_app.R- Script to run and control ubiquity ShinyApp -
server.R- Server script -
ui.R- UI script
Running the model
Next simply open ubiquity_app.R in RStudio and source
it, and RStudio will open in a window associated with that application.
The ShinyApp tends to work better when run within a browser.
After starting the App will load the default dosing from the file and run the model. The first model output will be plotted. And as you make changes to the App, those changes will be reflected in the user log at the bottom of the screen. You can change the outputs being plotted, the axis limits, and other aspects of the plot using the plot controls below the figure.
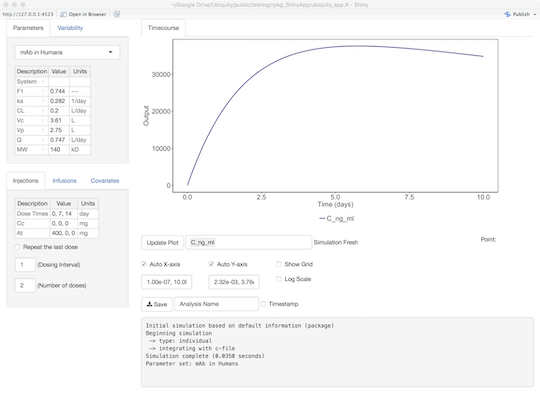
The intent is to provide a quick method to get the model up and running with default behaviors that will provide for most of what users will need. The behavior of the ShinyApp can be further customized (described below). First we will discuss the default interface:
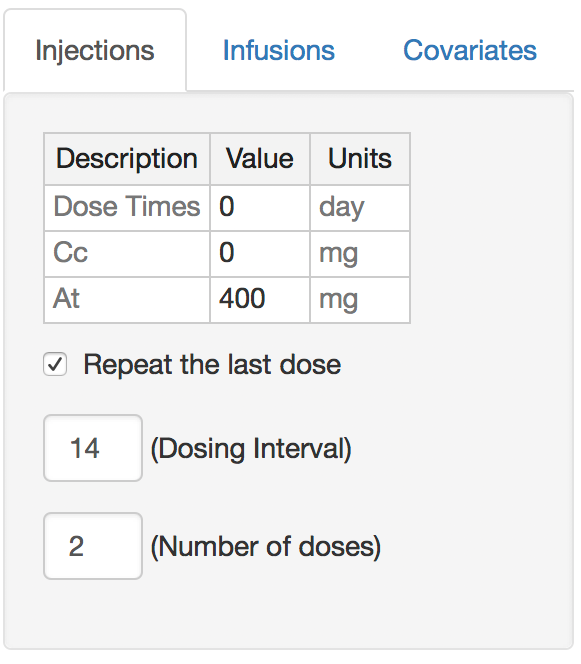
Altering dosing
A list of bolus times and values can be specified as comma-separated values. To dose in a repeated fashion check the ``Repeat the last dose’’ box, provide a dosing interval (in the same units as the dosing times), and give the number of doses to administer. The example here will repeat the 400 mg dose every two weeks for two doses.
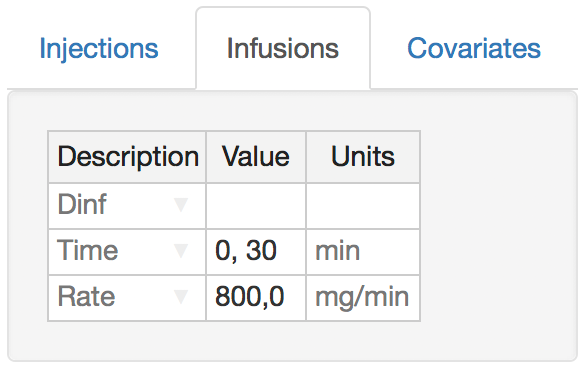
Rates of infusion are specified as a set of switching times and corresponding infusion rates (both separated by commas). The infusion rates are held constant until the next switching time. The example here would infuse the drug at 800mg/min starting at time 0, and the infusion will be turned off (set to 0) after 30 minutes.
Population simulations
If the variability for system parameters has been specified
(<IIV:?>? and <IIVCOR:?>?) then a
“Variability” tab will be visible. Simply check the “Simulate with
variability” box, alter the number of subjects, prediction interval, and
elements of the variance-covariance matrix to simulate the desired
population response.
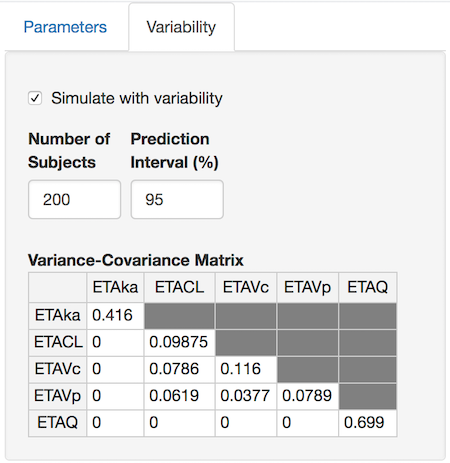
When you select “Update Plot”, each selected output will be plotted (mean – solid line, selected confidence interval – shaded region).
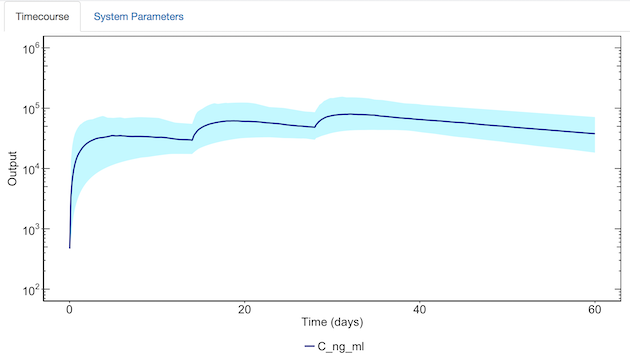
Saving model results
To save the model results you need to only provide a descriptive name for the analysis, push the save button, and click on the link that is generated. If you select the time stamp, this will be appended to the name of the zip file. By default the following files will be saved:
-
analysis_single_dose_400_mg_lib.r,analysis_single_dose_400_mg.r- These two files can be used to recreate the results from the ShinyApp. -
analysis_single_dose_400_mg_timecourse.png- Time-course with the simulation results -
analysis_single_dose_400_mg.csv- Predictions containing each timescale in the model, every state, and output -
analysis_single_dose_400_mg_RX.html- A file for each report (X) that has been specified inubiquity_app.R(See below) -
system.txt- The system file describing the model -
ubiquity_log.txt- The log file of what the user has done
Model scripts
The files analysis_NAME.r and
analysis_NAME_lib.r can be used in two ways. First, these
two files can be used as stand-alone files to recreate the simulation
results from the ShinyApp. Creating Simulation Scripts The main script
(analysis_NAME.r) can be used as a starting point to create
simulation scripts. To do this it is necessary to modify First comment
out the sourcing of the standalone library:
#source("analysis_NAME_lib.r");Next (with this script and the system.txt file)
uncommenting the following lines that will force the system to be
rebuilt and any changes to the system file loaded.
if(!require(ubiquity)){
source(file.path('library', 'r_general', 'ubiquity.R')) }
cfg = build_system(system_file="system.txt")Controlling what is saved
In certain instances you may not want to allow the user access to
everything that is saved by default. If you edit the
ubiquity_app.R you can alter what is being pushed to the
user by setting the relevant fields to FALSE.
cfg$gui$save$system_txt = TRUE
cfg$gui$save$user_log = TRUEModel documentation
Model diagram
It can be difficult for a user to look at a set of parameters, dosing inputs, and outputs and determine how a model works. To aid the user it can be useful to create a model diagram. To utilize the majority of the screen space, the image should be roughly twice as wide as it is tall.
You can create this image in your favorite drawing program. Inkscape is a free vector drawing program available at inkscape.org. If you run the following:
system_fetch_template(cfg, template="Model Diagram")it will create the file system.svg. You can edit it and
save it as a portable network graphics (png) file named
system.png in the directory with the
ubiquity_app.R file. When you start the ShinyApp it should
find that file and create a tab in the plotting area of the App.
Model reports
Sometimes you need more detail than an annotated model diagram. Or
you may need to perform some calculations based on the results of a
simulation. It may be better to present information in a tabulated
format. For these scenarios you can create an RMarkdown file. Create a
copy the report template (system_report.Rmd) and a script
to test it (test_system_report.r):
system_fetch_template(cfg, template="Shiny Rmd Report")When this is called you’ll have access to the ubiquity model object
(cfg) and the simulation results som. The
simulation results will take the form of a deterministic simulation (see
the output of ?run_simulation_ubiquity) or, if variability
is checked, a stochastic simulation (see output of
?simulate_subjects)
To tell the ShinyApp to load the report (named
system_report.Rmd), the following lines need to be added to
ubiquity_app.R.
cfg$gui$modelreport_files$R1$title = "Tab Title"
cfg$gui$modelreport_files$R1$file = "system_report.Rmd"The title field is the title of the tab in the ShinyApp
and file is the name of the file that you created. You can
create up to five reports this way, and these are differentiated by the
list name (R1, R2, … R5).
The file test_system_report.r can be used to debug your
model report file. First run your model in the ShinyApp with the reports
disabled. When the App runs it stores the state (cfg) and
the current simulation results (som) in the following data
files:
-
cfg\rightarrowtransient/rgui/default/gui_state.RData -
som\rightarrowtransient/rgui/default/gui_som.RData
Next load the test script, modify the report name accordingly, and run it:
load("transient/rgui/default/gui_som.RData")
load("transient/rgui/default/gui_state.RData")
params = list()
params$cfg = cfg
params$som = som
rmarkdown::render("system_report.Rmd",
params = params,
output_format = "html_document")This should generate your report in HTML format. You can insert debugging commands in the report and test it while you add information. Once you have it working, you can enable the report as described above and it should appear as a tab viewable by the user.
Not all reports may depend on user input. For example, if you have an
RMarkdown file that documents your modeling assumptions or the model
construction process then the content will not change as the user
interacts with the App. To use a pre-rendered report you simply have to
replace .Rmd with .html:
cfg$gui$modelreport_files$R1$title = "Tab Title"
cfg$gui$modelreport_files$R1$file = "system_report.html"This will tell the App to read in the html, prevent rendering the report, and speed things up for the user.
User definable functions
To customize the simulation and plotting within the ShinyApp it is
necessary to create custom functions. These need to be placed in a file.
For example these could be placed in mylibs.r in the main
template directory. Lastly, you need to tell the ShinyApp to load this
file. You need to edit ubiquity_app.R and add the following
line:
cfg$gui$functions$user_def = 'mylibs.r'Custom simulation commands
By default the ShinyApp will run simulations using
run_simulation_ubiquity (individual) or
simulate_subjects (population). These take as inputs the
parameters vector containing the current parameter set with
any changes made through the interface and cfg data
structure. You can create your own functions and overwrite these by
defining the following:
Individual Simulations
cfg$gui$functions$sim_ind = 'function_name(parameters, cfg)'Population Simulations
cfg$gui$functions$sim_var = 'function_name(parameters, cfg)'Note that in order to use the default plotting functionality these
functions need to return values in the same format as the default
functions for individual (run_simulation_ubiquity) and
population (simulate_subjects) simulations.
Custom plotting commands
To customize the plotting output you need to create functions that
can utilize the simulation output som. For individual and
population simulations this is the output of
run_simulation_ubiquity and simulate_subjects,
respectively. Unless user-specified simulation functions are being used.
In that case som can have any format needed. It is possible
to also access the parameters vector containing current
parameter set with any changes made through the interface and
cfg data structure. You can create your own plotting
functions and overwrite these by defining the following in
ubiquity_app.R:
Indiviudal Plot
cfg$gui$functions$plot_ind = 'function_name(cfg, parameters,som)'Population Plot
cfg$gui$functions$plot_var = 'function_name(cfg, parameters,som)'Deployment on a Shiny Server
To deploy the ShinyApp on a shiny server you need to indicate this by
modifying ubiquity_app.R and changing the deploying
variable to TRUE:
deploying = TRUEThen the ShinyApp needs to be initialized on the server. You can do
this two ways. First you can create an empty file called
REBUILD that needs to be deployed along side the other app
files. You can do this with the touch command from the
nat.utils package:
nat.utils::touch("REBUILD")This will force the initialization the first time the app is loaded.
This works well if you are using rsconnect to deploy your
app. Alternatively, you can manually sync the files and run the
following as the shiny user on the server.
If the App fails to run on the Shiny Server check the following:
- Make sure all of the correct packages have been installed and are available to the user the Shiny Server is running under.
- Make sure the ownership and permissions on the server files is correct.
To test deployment you can install the shinyVM